Molecular imaging of isolated peptidoglycans as novel platform for antibiotic development
Molecular imaging of a native isolated Escherichia coli peptidoglycan sacculus. Note the nanoscale resolution of single glycan chains, allowing the interpretation of murein network morphology on a molecular scale. (white and blue arrows mark peptidoglycan strands).
Molecular imaging of ampicillin-treated isolated Escherichia coli peptidoglycan sacculus. Note the nanoscale resolution of single structurally impaired peptidoglycanglycan strands, allowing the depiction of affected peptidoglycan morphology after ampicillin treatment (white arrow = altered peptidoglycan strand evoked by ampicillin treatment).
Natural products´ mechanisms of action
Although modern medicinal chemistry has achieved striking advances in the past, the plethora of new anti-infective and anti-cancer agents are still initially identified in a plant, animal or microorganism.
The subsequent elucidation of the mechanisms of action of such bioactive natural products (NPs) is yet still an essential but tedious process, hindered by a number of shortcomings.
In order to identify a target structure in an alternative way, our group is using ultrastructural morphological assessments of cells, tissues and whole organisms as a shortcut to pinpoint potential mechanisms of action of a bioactive NP. This information can subsequently allow the more detailed description of a possible mechanism of action as well as of protein targets affected by the investigated NP.
(Ultra)-structural elucidation by microscopic techniques
The assessment of relevant structural alterations in our group is performed by a variety of microscopic techniques. On the one hand, classical light microscopic approaches (brightfield, darkground, phase contrast, polarization, fluorescence as well as DIC) are represented in our lab, allowing the examination of biological material on the lower micrometer scale (lateral resolution down to 0.2 µm).
On the other hand, innovative investigations on a nanometer scale are also possible by four atomic force microscopes (AFMs) in our lab, allowing lateral resolutions down to a fraction of an ångström (typically down to roughly 5 nm in biological material and under ambient conditions).
As a topographical technique, gaining insight into internal features of biological material by atomic force microscopy is still a challenging process. We therefore established an ultra-microtomic protocol, allowing insight into internal features of virtually any cell, tissue and organism by AFM for the first time. This technique was used in the recent years to describe the mechanism of action of different bioactive natural products.
Developed workflow for the facile ultrastructural elucidation of internal features of organisms, tissues or single cells by Atomic Force Microscopy (Herrmann et al., J. Struc. Biol. 2019)
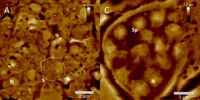
AFM-elucidated ultrastructure of C. elegans L4 hermaphrodite´s spermathecal tissues after treatment with the quassinoid ailanthone (C, 50 µM) and compared to untreated morphology (A). S = single spermatocyte, Sp = single spermatid. (Adapted from Knetzger et al., Doi:10.3390/molecules26237354)
In silico and enzymatic techniques
Additionally, our group is also focused on enzymatic as well as in silico techniques. After the identification and structural elucidation of a target protein, in silico techniques can be applied to screen large substance databases for compounds potentially interacting with the protein of interest (virtual screening). Furthermore, molecular docking analyses subsequently allow the estimation of the possible interaction of a set of given compounds with a protein, enabling us to identify the most promising hits for in vitro testing.
Spectroscopy-based in vitro inhibition experiments employing the recombinantly produced target protein are afterwards applied in order to verify the in silico predictions as well as to gain further insight into the mechanism of action of an enzyme inhibitor.
Available methods in the Herrman lab
- Atomic Force Microscopy (altogether 4 Bio-AFMs available)
- Sample preparation by ultra-microtomy
- Light microscopy (epifluorescence, phase contrast, DIC...)
- In silico drug design (pharmacophore elucidation, virtual screening, molecular docking)
- Recombinant expression of target proteins
- Assessment of enzyme kinetics
Current research projects
- AFM-based molecular analysis of isolated peptidoglycan sacculi to establish a screening assay for novel cell wall inhibiting antibiotics
- Ultrastructural evaluation of the antibacterial effects of galectins 3 and 4 against E. coli (cooperation with USP Sao Paulo, Prof. Dr. Baruffi)
- Ultrastructural evaluation of the mechanism of action of different quassinoids and proanthocyanidins against Caenorhabditis elegans
- Ultrastructural evaluation of anthelmintic effects exerted on Caenorhabditis elegans by treatment with a proanthocyanidin-rich extract from Combretum mucronatum
- Development of ultramicrotomy-based techniques allowing the ultrastructural characterization of cells, tissues and organisms by AFM and light microscopy
Supervised PhD theses
- Daniel Amiteye, "In silico identification and AFM-based ultrastructural evaluation of natural products as inhibitors of the bacterial cell wall synthesis", since 2021
Supervised master theses
- Niklas Mayer, 2024
- Mirko Hirsch, 2023
- Leonardo Elsbroek, "Ultrastructural characterization of isolated peptidoglycan sacculi by atomic force microscopy", 2021/22
- Nicola Knetzger, "Elucidating the mechanism of action of two quassinoids against Caenorhabditis elegans by Atomic Force Microscopy (AFM)", 2020
- Daniel Dornbusch, "Entwicklung einer rasterkraftmikroskopischen Methode zur Visualisierung morphologischer Veränderungen an Leishmanien nach Behandlung mit Tubulin-modifizierenden Wirkstoffen", 2019
- Katharina Possart, "Rationale Suche nach Naturstoffen mit antitrypanosomaler Aktivität", 2019
- Nirina Sivakumar, "Naturstoffe mit Hemmwirkung auf die Pteridinreduktase 1 von Leishmania major und Trypanosoma brucei", 2016
- Alexandra Heithorst, "Mechanistische Untersuchungen zur Hemmung trypanosomaler Enzyme durch ausgewählte Naturstoffe", 2016